- April 2021
- Volume 27
- Issue 3
Genomic Cancer Profiling: Setting a New Standard in Lung Cancer Treatment

Genomic profiling has revolutionized the way cancer care is being approached. Following the completion of the Human Genome Project in 2003, which provided the DNA reference blueprint, many actionable genomic alterations have been identified.
Non-small cell lung cancer (NSCLC) is a genetically diverse disease, with at least 9 genetic alterations that are clinically accessible for targeted therapy. Some of these are virtually exclusive to NSCLC, such as EGFR mutations. Others span a broad spectrum of cancer types, like BRAF and NTRK alterations.1 Additional biomarkers, including PD-L1, mismatch repair deficiency or microsatellite instability, and tumor mutational burden are FDA-approved markers of immunotherapy, a third therapeutic pillar of NSCLC beyond targeted therapy and conventional chemotherapy.2
With the breadth of targetable mutations, next-generation sequencing provides a highly sensitive, parallel approach to identifying the complete set of relevant biomarkers to guide therapy, allowing clinicians to match biology with the appropriate therapy and refining the one-size-fits-all approaches previously applied in NSCLC.3
Results of genomic testing are a critical component of the diagnosis, complementing previous histology-based assessments, according to key national cancer treatment guidelines, including those published by the American Society of Clinical Oncology and National Comprehensive Cancer Network.4 These complete results guide critical therapy decisions and, ideally, they are available prior to initiation of any systemic therapy.
The number of actionable mutations in NSCLC is ever-expanding, and approvals to target alterations in KRAS, HER2/neu and NRG1/HER3 are expected in the near future.5,6 Most nonsquamous NSCLC diagnoses will be characterized by the presence of a driver mutation. Therefore, the yield and effect of genomic testing will continue to grow and improve outcomes in NSCLC.
The nature of genomic alterations is complex and diverse. It can include simple alterations, such as point mutations in EGFR or NTRK; or higher genomic rearrangements, like splicing variants in the MET gene or fusion products of 2 seemingly unrelated genes. Gene fusion is the joining of 2 genes, and when the binding partner upstream of the actual gene is responsible for activation of a typically dormant gene it results in malignant transformation. One of these genomic alterations is the NTRK gene fusion product. Fusion of 1 of the NTRK genes to an activating promoter gene can activate NTRK-related signaling in NSCLC.7-9 As with other oncogenes, no clinical or histological characteristic hints at the presence of an NTRK fusion, and agnostic testing of all patients with NSCLC is critical to identify the mutation.
Several challenges have made stringent genomic testing for all patients with NSCLC difficult, including lack of sufficient biopsy specimen, viability of biopsy specimen, tissue utilization during histological work up, insurance coverage, and delays in turnaround time for laboratory results when there is a clinical urgency to initiate systemic therapy.10 Although the challenges are clearly affecting real-world care delivery for lung cancer, the following case is a good example of why it is worthwhile, even in later lines of therapy.
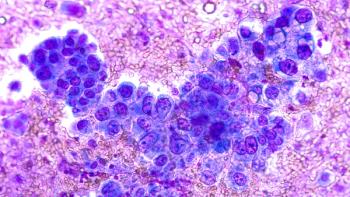
A 41-year-old female patient received a diagnosis of adenocarcinoma of the lung after presenting with shortness of breath. Her biopsy revealed a PD-L1 level of 80% and no actionable mutation was seen on liquid biopsy. No additional tissue was available for genomic testing. The patient was initiated on single-agent immunotherapy and showed disease progression on the first follow-up scans. Chemotherapy was then added to immunotherapy, stabilizing her disease for 3 months prior to clinical progression. A repeat tissue biopsy was obtained and retested on an RNA-based
tissue platform. An NTRK3 fusion was identified, and the patient was started on targeted therapy with near complete response on the first follow-up scan (Figure).
While liquid biopsy is a convenient and ubiquitously available testing modality for the detection of point mutations and small deletions, fusions and other more complex genomic alterations are more sensitively detected on tissue-based assays. A negative evaluation by liquid biopsy should be followed by a tissue-based assay, and a negative tissue-based evaluation on a DNA platform should be complemented by an RNA-based tissue analysis. This effort of comprehensive tissue testing maximizes the probability of oncogene detection, facilitates the selection of appropriate therapies, and helps avoid ineffective therapies and their associated medical and financial toxicities.
Improved outcomes following genomically guided therapy have been extensively demonstrated, and a reduction in adverse effects has demonstrated superior quality of life and significant cost savings for patient care delivery.11,12 From a patient perspective, it is an improvement in quality and quantity of life, with synchronous reduction in cost of care delivery.
Or as one oncologist put it, genomic testing assures “the right therapy for the right patient at the right time.”
AUTHOR INFORMATION
Martin F. Dietrich, MD, PhD, is a medical oncologist at Florida Cancer Specialists & Research Institute and an assistant professor of internal medicine at the University of Central Florida in Orlando. He is a board-certified internist and medical oncologist with a special research interest interest in lung and breast cancer treatment, as well as genetic evaluation and counseling of somatic and hereditary syndromes.
REFERENCES
1. Targeted drug therapy for non-small cell lung cancer. American Cancer Society. Updated February 16, 2021. Accessed February 23, 2021. cancer.org/cancer/lung-cancer/treating-non-small-cell/targeted-therapies.html
2. Huang D, Zhang F, Tao H, et al. Tumor mutation burden as a potential biomarker for PD-1/PD-L1 inhibition in advanced non-small cell lung cancer. Target Oncol. 2020;15(1):93-100. doi:10.1007/s11523-020-00703-3
3. Coco S, Truini A, Vanni I, et al. Next generation sequencing in non-small
cell lung cancer: new avenues toward the personalized medicine. Curr Drug Targets. 2015;16(1):47-59. doi:10.2174/1389450116666141210094640
4. Hechtman JF, Benayed R, Hyman DM, et al. Pan-trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol.¡2017;41(11):1547-1551. doi:10.1097/PAS.0000000000000911
5. Bernicker EH, Allen TC, Cagle PT. Update on emerging biomarkers in lung cancer. J Thorac Dis. 2019;11(1):S81-S88. doi:10.21037/jtd.2019.01.46
6. Schram A, O’Reilly E, Somwar R, et al. Clinical proof of concept for MCLA-128, a bispecific HER2/3 antibody therapy, in NRG1 fusion-positive cancers. Presented at: 2019 AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics; October 26-30, 2019; Boston, MA. Abstract PR02.
7. Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov.¡2015;5(1):25-34. doi:10.1158/2159-8290.CD-14-0765
8. Amatu A, Sartore-Bianchi A, Siena S.¡NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1(2):e000023. doi:10.1136/esmoopen-2015-000023
9. Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 2015;7:129.¡doi:10.1186/s13073-015-0252-1
10. Gutierrez ME, Choi K, Lanman RB, et al. Genomic profiling of advanced non-small cell lung cancer in community settings: gaps and opportunities. Clin Lung Cancer. 2017;18(6):651-659. doi:10.1016/j.cllc.2017.04.004
11. Oizumi S, Kobayashi K, Inoue A, et al. Quality of life with gefitinib in patients with EGFR-mutated non-small cell lung cancer: quality of life analysis of North East Japan Study Group 002 Trial. Oncologist. 2012;17(6):863-870. doi:10.1634/theoncologist.2011-0426
12. Seo MK, Cairns J. Do cancer biomarkers make targeted therapies cost-effective? A systematic review in metastatic colorectal cancer. PLoS One. 2018;13(9):e0204496. doi:10.1371/journal.pone.0204496
Articles in this issue
almost 5 years ago
Path to Better Payment System to Be Clear Over Timealmost 5 years ago
AJMCtv Interviews: April 2021almost 5 years ago
Medical World News: Clinical, Regulatory, and Managed Care Updatesalmost 5 years ago
Conference Coverage: Association of Community Cancer Centers 2021almost 5 years ago
Disparities in Cancer Care: Has Precision Medicine Widened the Gap?almost 5 years ago
The Impact of Reinsurance of Gene Therapies on Employer Financial RiskNewsletter
Stay ahead of policy, cost, and value—subscribe to AJMC for expert insights at the intersection of clinical care and health economics.